Beschreibung
In this subproject, the properties of enzymes under process conditions is modelled in atomistic detail by molecular dynamics simulations and combined with thermodynamic modelling of substrate-solvent mixtures and with kinetic modelling of biocatalytic processes. The integration of molecular, thermodynamic, and kinetic modeling provides tools for the rational design of enzymes and the for process engineering.
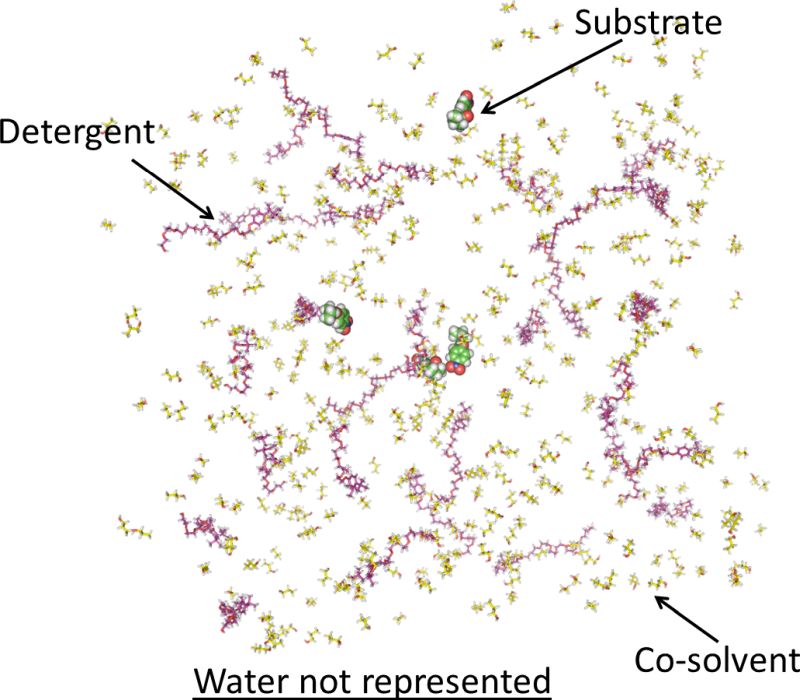
Biocatalytic reactions are usually not performed in pure water, but in complex mixtures which contain a substrate concentration close to its solubility limit, co-solvents, and detergents in water or organic solvents (Fig.1).
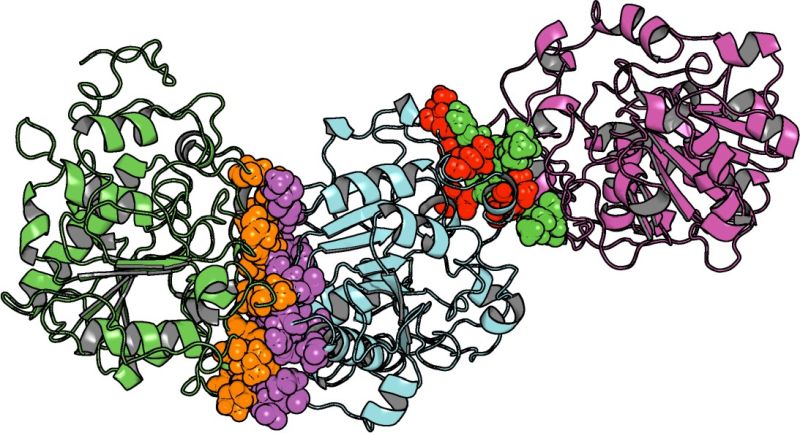
The components compete for binding to the protein surface and to the active site of the enzyme. Moreover, the substrate-solvent mixture might also affect aggregation of the protein which might lead to inactivation (Fig. 2).
 |
|
 |
|
Enzyme function is a process that occurs on a wide range of time scales, from atomistic motions in the ps and ns time scale to substrate turnover rates in the ms to s time scale. The molecular dynamics simulation of an enzyme in realistic substrate-solvent mixtures is a first principle approach to study molecular structure and conformational dynamics of the protein, its interaction with substrates, solvents, and additives, as well as transport properties and thermodynamics of the substrate-solvent mixture.
As a model system, we study the binding of substrates and of organic solvent molecules by molecular dynamics simulations on the μs time scale to derive binding constant from first principles (Video). Alternatively, kinetic parameters are derived from kinetic modeling of biocatalytic experiments using parameter estimation. Scale bridging between the ms time scale of molecular dynamics simulations and the s time scale of biocatalytic experiments is achieved by relating the parameters derived from molecular simulation to kinetic modeling. The simulation results can also be applied to solvent selection, which is relevant for stability of the enzyme, product recovery, and substrate solubility. By performing molecular dynamics simulation of realistic environment it is possible to calculate, by thermodynamic integration or free energy perturbation, the thermodynamic activity of substrate and product molecules in different environment, discriminating between solvent and biocatalysts interactions. Noteworthy, it is possible to simulate complex mixture with several components. The possibilities to calculate such data with high precision represents a powerful tool for solvent design and selection.
VIDEO
The lipase B from Candida antarctica was simulated in an aqueous environment with substrate (pNP-butyrate) at a concentration of 1 mM. Substrates molecules bind to the protein surface and subsequently enter the substrate access channel which leads to the active site.

