| Abstract |
Atoms and molecules are the constituents of matter, be it inorganic or organic, non-biological
or biological. Thus, all physical properties of matter can in principle be understood by analyzing
these fundamental constituents and their mutual interactions. Even though the hard sphere model of
atoms is a great simplification of physics, it is often used to visualize and analyze molecular
structures as well as fluids and solids. In this talk I will present methods that we contributed to
the field of molecular visualization and analysis, based on the hard sphere model, over the last
few years. Most of these methods are applicable to large atomic systems both in terms of
visualization and analysis due to the exploitation of GPU-based rendering and computation.
The major part of the talk concerns molecule-molecule interactions, the transport of small
molecules into cavities of large molecules, and the dynamic behavior of cavities in molecular
trajectories. To analyze the potential interaction surface of macromolecules with other molecules
like ligands, the solvent excluded surface (SES) is most often used. Methods for real-time
rendering of the SES suitable for molecular trajectories of even large proteins will be presented
along with the definition and computation of the generalization of the SES, called the ligand
excluded surface (LES). For the computation of cavities, we utilize the concept of the Voronoi
diagram of spheres. We show that it can be efficiently used to compute both static and dynamic
molecular paths. Furthermore, I will present visualizations that we developed based on this concept
to allow the interactive analysis of the computed cavities, which often is required to incorporate
the knowledge of expert users.
In the rest of the talk I will show that by exploiting the fact that very large macromolecular
systems often consist of many recurring structures, we can greatly speed up the rendering and
achieve interactive frame rates for molecular structures with more than a billion atoms. By using
deferred shading, we can obtain a smooth transition between the atomic details for close-ups and
surface-like structures for objects far away from the observer without changing the underlying
representation.
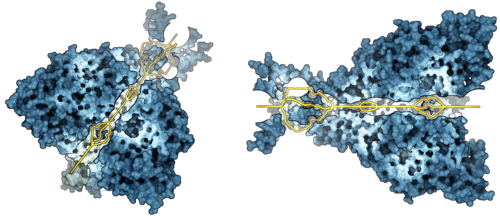
Figure: Molecular path visualization of a sodium ion channel (PDB: 3HGC). The
channel is important for epidermal ion transport into the cell.
|
